Project objectives
The main objectives of the project are: (i) to study a series of vector molecules for targeted drug delivery and to outline the most prospective candidates via rational molecular design; (ii) to investigate the mechanism of targeting of these (bio)molecules to model cell membranes representing certain cell types and of their receptor recognition therein. The specified problems are addressed by multiscale molecular modelling – employing methods from classical molecular dynamics to electron-correlated DFT. An important educational goal is the training of early-stage researchers.
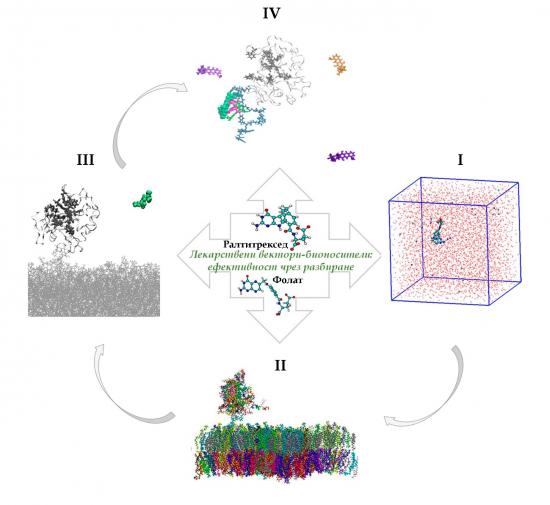
Schematic representation of the key stages of the project: (I) description of the molecular structure of the vectors in saline; (II) development of a suitable model membrane with an embedded receptor; (III) elucidation of the targeting molecules binding to the receptor; (IV) directed transport of bioactive cargo and nomination of the most promising vectors (raltitrexed and folate)
Research on the project
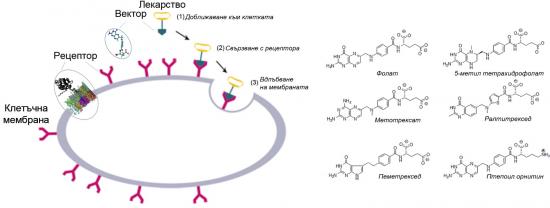
The series of vector molecules includes folate, 5-methyl tetrahydrofolate, raltitrexed, pemetrexed, methotrexate, and pteroyl ornithine in saline.
(left) Illustration of the modelled part of receptor-embedded cell membrane and (right) chemical structures of the studied vector ligands
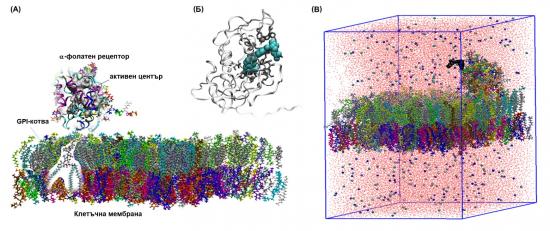
For the purpose of studying the interaction of the targeting ligands with the folate receptor-a, a model neoplastic cell membrane is constructed, based massively on experimental data. It consists of a lipid bilayer mimicking a lipid raft built of 370 lipids of 35 different types with asymmetric charge and an embedded molecule of FRa. The size of the simulated models is ~185000 atoms.
Pictorial representation (on actual snapshots) of (А) the model cell membrane with an embedded receptor molecule, (B) the crystallographic structure of the complex folate/folate receptor-a (PDB ID: 4LRH; DOI: 10.1038/nature12327), and (C) the simulated systems membrane/receptor/vector ligand in saline
The process of binding of each of these molecules to a target receptor: the folate receptor-a (FRa), a protein overexpressed on the surface of cancer cells, is monitored.
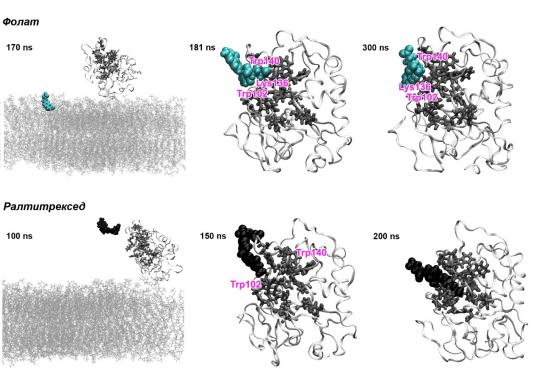
Illustration of the spontaneous binding of (top) folate and (bottom) raltitrexed to the folate receptor-a and positioning of the ligands in the binding pocket; membrane lipids are depicted as grey lines, the protein – as white ribbons, the amino acids from the active site – as dark grey licorice, and the targeting ligands – as cyan (folate) or black (raltitrexed) spheres
Outcome
Spontaneous binding at the nanosecond timescale of each of the ligands with the receptor is observed by atomistic molecular dynamics. However, only folate, raltitrexed, and 5-methyl tetrahydrofolate recognize the protein active centre. It is established that in order to achieve such selective binding the ligand should be able to take part simultaneously in a set of specific interactions with FRa: p-stacking, hydrogen bonding, van der Waals and electrostatic attraction. The key amino acids for ligand recognition are identified. It is revealed that the ligand-receptor complex formation process involves several steps, one of them requiring reorganization of the active site, and the initial binding pose is observed for the first time. The differences in the affinities of the ligands for the receptor known experimentally are explained. The four most prospective vector molecules (folate, raltitrexed, pemetrexed, and 5-methyl tetrahydrofolate) are included into model systems for targeted drug delivery, for which the interaction with the receptor is tracked. It turns out that folate and raltitrexed manage to bind the cargo conjugated to them close to the protein active site whereas the other two ligands lose part of their functionality.
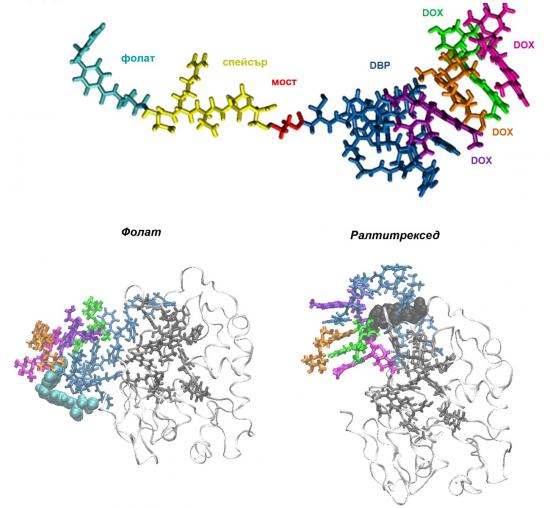
Structure of (top) the system for targeted drug delivery containing folate with an attached complex of four molecules of the chemotherapeutic doxorubicin (DOX) adsorbed on a drug-binding peptide (DBP) and (bottom) this system with two different targeting vectors bound to the folate receptor-a; the protein is depicted as white ribbons, the amino acids from the active site – as dark grey, DBP – as dark blue, DOX – as various colors licorice, and the targeting ligands – as cyan (folate) or black (raltitrexed) spheres
Therefore, raltitrexed and folate are suggested as the most suitable targeting units.
Several students and young researchers are trained within the framework of the project.
Impact
The new knowledge may be employed mainly for development of systems for targeted drug delivery based on folate and its derivatives. The obtained results may be used for optimization of the constitution and enhancement of the efficiency of such assemblies for active drug transport. The clarified mechanism of the binding process of ligands to FRa enables rational design of new folate derivatives, which could serve as receptor agonists or antagonists. The constructed model cell membrane may be applied in a broad range of biomolecular simulations aimed at combatting malignant neoplasms.
Synopsis
Two vector molecules are suggested, which recognize efficiently the folate receptor-a and bind to it. They may be included in different modern active drug delivery formulations. A set of molecular-level guidelines is derived, describing the way of binding of bioactive cargo in drug delivery systems based on folate or its antimetabolites. Molecular details about the binding of the series of vector molecules to the receptor are deciphered, which allows more focused search of similar compounds that could activate or deactivate the macromolecule. This could lead to enhancement or inhibition of specific biological processes related to the presence of folate or its derivatives in living cells.
The obtained results are summarized in several scientific papers and presented at various events.